mBuild Compound Hierarchy
mBuild Compound hierarchy
mBuild allows for the hierarchical construction of systems from smaller Compounds. There is no rigid hierarchy in mBuild aside from the lowest level of the hierarchy representing a point particle. That is, a system can be composed of any arbitrary number of levels of Compounds. While very convenient for constructing complex systems from simpler/smaller components, it can make it challenging to fully decode the structure of a given Compound, especially if one wishes to reference/modify specific parts of a Compound.
Let us start be examining ethane, a simple test Compound provided in the mBuild molecule library.
import mbuild as mb
from mbuild.lib.molecules.ethane import Ethane
ethane = Ethane()
There are many ways to look at what is contained within a Compound. For example, we can look at the lowest level in the sub-Compounds by calling the particles function.
for particle in ethane.particles(): print(particle)
<C pos=([ 2.5032e-17 -1.4000e-01 2.1906e-18]), 4 bonds, id: 4421537024>
<H pos=([-1.0700e-01 -1.4000e-01 1.3944e-17]), 1 bonds, id: 4421679760>
<H pos=([ 0.0357 -0.2169 0.0653]), 1 bonds, id: 4419309280>
<H pos=([ 0.0357 -0.1581 -0.0993]), 1 bonds, id: 5397016048>
<C pos=([0. 0. 0.]), 4 bonds, id: 5396937120>
<H pos=([0.107 0. 0. ]), 1 bonds, id: 4419175568>
<H pos=([-0.0357 0.0769 0.0653]), 1 bonds, id: 5397035136>
<H pos=([-0.0357 0.0181 -0.0993]), 1 bonds, id: 5388459072>
Looping over the particles tells us what particles are contained in the system, but not the actual structure of the hierarchy.
To begin to look at the hierarchy, we could loop over the children. Here we see that in the next level, we have two CH3 compounds, each containing 4 of the 8 particles.
for child in ethane.children: print(child)
<CH3 4 particles, 3 bonds, non-periodic, id: 4419177776>
<CH3 4 particles, 3 bonds, non-periodic, id: 5397036720>
Of course, this doesn’t provide us with a full view of the hierarchy, as we still need to explore the structure of the CH3 Compounds. For example, let us simply embed a second loop in the code above, looping over the children of each CH3:
for CH3 in ethane.children:
print(CH3)
for child in CH3.children: print("-", child)
<CH3 4 particles, 3 bonds, non-periodic, id: 4419177776>
- <C pos=([ 2.5032e-17 -1.4000e-01 2.1906e-18]), 4 bonds, id: 4421537024>
- <H pos=([-1.0700e-01 -1.4000e-01 1.3944e-17]), 1 bonds, id: 4421679760>
- <H pos=([ 0.0357 -0.2169 0.0653]), 1 bonds, id: 4419309280>
- <H pos=([ 0.0357 -0.1581 -0.0993]), 1 bonds, id: 5397016048>
<CH3 4 particles, 3 bonds, non-periodic, id: 5397036720>
- <C pos=([0. 0. 0.]), 4 bonds, id: 5396937120>
- <H pos=([0.107 0. 0. ]), 1 bonds, id: 4419175568>
- <H pos=([-0.0357 0.0769 0.0653]), 1 bonds, id: 5397035136>
- <H pos=([-0.0357 0.0181 -0.0993]), 1 bonds, id: 5388459072>
Note that particles, by definition, are the lowest level in the hierarchy and thus do not have children.
mBuild does provide the successors function to automatically loop over all levels, however, for complex and/or large molecules, the output can be challenging to parse.
for successor in ethane.successors(): print(successor)
<CH3 4 particles, 3 bonds, non-periodic, id: 4419177776>
<C pos=([ 2.5032e-17 -1.4000e-01 2.1906e-18]), 4 bonds, id: 4421537024>
<H pos=([-1.0700e-01 -1.4000e-01 1.3944e-17]), 1 bonds, id: 4421679760>
<H pos=([ 0.0357 -0.2169 0.0653]), 1 bonds, id: 4419309280>
<H pos=([ 0.0357 -0.1581 -0.0993]), 1 bonds, id: 5397016048>
<CH3 4 particles, 3 bonds, non-periodic, id: 5397036720>
<C pos=([0. 0. 0.]), 4 bonds, id: 5396937120>
<H pos=([0.107 0. 0. ]), 1 bonds, id: 4419175568>
<H pos=([-0.0357 0.0769 0.0653]), 1 bonds, id: 5397035136>
<H pos=([-0.0357 0.0181 -0.0993]), 1 bonds, id: 5388459072>
Making it easier to examine Compound hierarchy
To make it easier to examine the structure, I recently added a function called print_hierarchy. This uses the treelib python library to handle formatting the output.
By default the function will looking for matching Compounds and group them to create a more compact representation.
ethane.print_hierarchy()
Ethane, 8 particles, 7 bonds, 2 children
└── [CH3 x 2], 4 particles, 3 bonds, 4 children
├── [C x 1], 1 particles, 4 bonds, 0 children
└── [H x 3], 1 particles, 1 bonds, 0 children
<treelib.tree.Tree at 0x141d1fb80>
Compounds are considered to be the same if they have:
- the same name attribute,
- same number of children,
- same number of particles,
- same number of bonds,
- the name attribute of children and name of the particles in children are the all the same and in an identical order
This will ensure that any Compound that is “cloned” (i.e., copied and unchanged) will be considered matching. However, if two Compounds contain the same particles (and ostensibly represent the same chemistry) but have different hierarchies, each will be considered unique. This is by design, because the goal is to be able to understand the hierarchy so that, if needed, one can reference and modify the correct sub-Compounds.
We can print the entire hierarchy, showing all duplicates if we wish.
ethane.print_hierarchy(print_full=True)
Ethane, 8 particles, 7 bonds, 2 children
├── [CH3]: 4 particles, 3 bonds, 4 children
│ ├── [C]: 1 particles, 4 bonds, 0 children
│ ├── [H]: 1 particles, 1 bonds, 0 children
│ ├── [H]: 1 particles, 1 bonds, 0 children
│ └── [H]: 1 particles, 1 bonds, 0 children
└── [CH3]: 4 particles, 3 bonds, 4 children
├── [C]: 1 particles, 4 bonds, 0 children
├── [H]: 1 particles, 1 bonds, 0 children
├── [H]: 1 particles, 1 bonds, 0 children
└── [H]: 1 particles, 1 bonds, 0 children
<treelib.tree.Tree at 0x141d1f760>
We can also index into a specific branch of the hierarchy:
ethane.print_hierarchy(print_full=True, index=1)
Ethane, 8 particles, 7 bonds, 2 children
└── [CH3]: 4 particles, 3 bonds, 4 children
├── [C]: 1 particles, 4 bonds, 0 children
├── [H]: 1 particles, 1 bonds, 0 children
├── [H]: 1 particles, 1 bonds, 0 children
└── [H]: 1 particles, 1 bonds, 0 children
<treelib.tree.Tree at 0x141ceb520>
The print_hierarchy function returns the treelib Tree, providing a user with increased functionality. For example,
tree = ethane.print_hierarchy(print_full=True, show_tree=False)
tree.depth()
2
tree.show(line_type="ascii-em")
Ethane, 8 particles, 7 bonds, 2 children
╠══ [CH3]: 4 particles, 3 bonds, 4 children
║ ╠══ [C]: 1 particles, 4 bonds, 0 children
║ ╠══ [H]: 1 particles, 1 bonds, 0 children
║ ╠══ [H]: 1 particles, 1 bonds, 0 children
║ ╚══ [H]: 1 particles, 1 bonds, 0 children
╚══ [CH3]: 4 particles, 3 bonds, 4 children
╠══ [C]: 1 particles, 4 bonds, 0 children
╠══ [H]: 1 particles, 1 bonds, 0 children
╠══ [H]: 1 particles, 1 bonds, 0 children
╚══ [H]: 1 particles, 1 bonds, 0 children
tree.save2file('tree.txt') #note, this appends to a file if it already exists
!cat tree.txt
Ethane, 8 particles, 7 bonds, 2 children
├── [CH3]: 4 particles, 3 bonds, 4 children
│ ├── [C]: 1 particles, 4 bonds, 0 children
│ ├── [H]: 1 particles, 1 bonds, 0 children
│ ├── [H]: 1 particles, 1 bonds, 0 children
│ └── [H]: 1 particles, 1 bonds, 0 children
└── [CH3]: 4 particles, 3 bonds, 4 children
├── [C]: 1 particles, 4 bonds, 0 children
├── [H]: 1 particles, 1 bonds, 0 children
├── [H]: 1 particles, 1 bonds, 0 children
└── [H]: 1 particles, 1 bonds, 0 children
print(tree.to_json(with_data=False))
{"Ethane, 8 particles, 7 bonds, 2 children": {"children": [{"[CH3]: 4 particles, 3 bonds, 4 children": {"children": ["[C]: 1 particles, 4 bonds, 0 children", "[H]: 1 particles, 1 bonds, 0 children", "[H]: 1 particles, 1 bonds, 0 children", "[H]: 1 particles, 1 bonds, 0 children"]}}, {"[CH3]: 4 particles, 3 bonds, 4 children": {"children": ["[C]: 1 particles, 4 bonds, 0 children", "[H]: 1 particles, 1 bonds, 0 children", "[H]: 1 particles, 1 bonds, 0 children", "[H]: 1 particles, 1 bonds, 0 children"]}}]}}
mBuild includes a function to flatten a Compound. This function essentially reduces the underlying hierarchy to simply represent the particles. This change is easy to view using the print_hierarchy function.
ethane_flat = mb.clone(ethane)
ethane_flat.flatten()
ethane_flat.print_hierarchy()
Ethane, 8 particles, 7 bonds, 8 children
├── [C x 2], 1 particles, 4 bonds, 0 children
└── [H x 6], 1 particles, 1 bonds, 0 children
<treelib.tree.Tree at 0x141d7b6a0>
Let us consider a more complex example that now is composed of two different molecule types.
from mbuild.lib.molecules.methane import Methane
system = mb.Compound(name="System")
system.add(Ethane())
system.add(Ethane())
system.add(Methane())
system.add(Methane())
system.add(Methane())
system.print_hierarchy()
System, 31 particles, 26 bonds, 5 children
├── [Ethane x 2], 8 particles, 7 bonds, 2 children
│ └── [CH3 x 2], 4 particles, 3 bonds, 4 children
│ ├── [C x 1], 1 particles, 4 bonds, 0 children
│ └── [H x 3], 1 particles, 1 bonds, 0 children
└── [Methane x 3], 5 particles, 4 bonds, 5 children
├── [C x 1], 1 particles, 4 bonds, 0 children
└── [H x 4], 1 particles, 1 bonds, 0 children
<treelib.tree.Tree at 0x141ceb250>
What happens if we have ports in a Compound? Let us examine printing the hierarchy of a CH2 moiety with 2 ports.
from mbuild.lib.moieties.ch2 import CH2
ch2 = CH2()
ch2.print_hierarchy()
CH2, 3 particles, 2 bonds, 5 children
├── [C x 1], 1 particles, 2 bonds, 0 children
├── [H x 2], 1 particles, 1 bonds, 0 children
└── [Port x 2], 0 particles, 0 bonds, 2 children
└── [subport x 2], 0 particles, 0 bonds, 4 children
└── [G x 4], 1 particles, 0 bonds, 0 children
<treelib.tree.Tree at 0x107694b20>
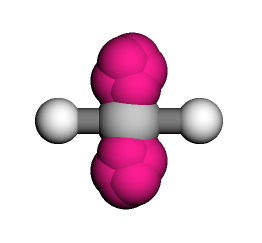
Here, points are essentially treated as if they were any other Compound, where the lowest level, G, corresponds to the 8 points which define the location and orientation of a port; these correspond to the pink blobs if we were to visualize the compound.
ch2.visualize(show_ports=True)